What Direction Does Rna Polymerase Read Dna
ii.1: Overview of Transcription
- Page ID
- 32270
Learning Objectives
- Identify the cardinal steps of transcription, the function of the promoter and the role of RNA polymerase.
- Distinguish between coding (RNA-like) and non-coding (template) strands of DNA. Empathise that within a single slice of Dna, either strand can be used as the template for different genes, but the RNA will withal be produced from five' → 3'.
- Depict a line diagram showing a segment of Dna from a gene and its RNA transcript, indicating which Dna strand is the template, the direction of transcription and the polarities of all DNA and RNA strands.
- Give examples of non-coding RNA molecules.
What is transcription?
Consider that all of the cells in a multicellular organism have arisen past division from a single fertilized egg and therefore, all have the same Deoxyribonucleic acid. Division of that original fertilized egg produces, in the example of humans, over a trillion cells, by the time a baby is produced from that egg (that's a lot of Dna replication!). Still, we likewise know that a baby is not a giant ball of a trillion identical cells, only has the many dissimilar kinds of cells that make up tissues like skin and muscle and bone and nerves. How did cells that have identical Dna turn out so different?
The answer lies in gene expression, which is the process past which the data in Dna is used. Although all the cells in a baby have the same DNA, each different prison cell type uses a dissimilar subset of the genes in that Deoxyribonucleic acid to direct the synthesis of a distinctive set of RNAs and proteins. The starting time step in gene expression is transcription, the process of copying information from DNA sequences into RNA sequences. This process is also known as Deoxyribonucleic acid-dependent RNA synthesis. When a sequence of DNA is transcribed, only one of the two Deoxyribonucleic acid strands is copied into RNA, when this RNA encodes a protein is information technology known as messenger RNA (mRNA).

Important features of transcription
- All RNA, mRNA every bit well as tRNA, rRNA, microRNA and more, is produced by transcription.
- Merely one strand of Deoxyribonucleic acid is used as a template by enzymes called RNA polymerases
- RNA is synthesized from 5' to 3'.
- RNA polymerases do not need primers to begin transcription.
- The iv ribonucleotide triphosphates (rNTPs) are ATP, GTP, UTP, and CTP.
- RNA polymerases begin transcription at DNA sequences called promoters.
- RNA polymerases stop transcription at sequences called terminators.
In transcription, an RNA polymerase uses only one strand of Dna, called the template strand, of a gene to catalyze synthesis of a complementary, antiparallel RNA strand. RNA polymerases apply ribose nucleotide triphosphate (NTP) precursors, in dissimilarity to Deoxyribonucleic acid polymerases, which apply deoxyribose nucleotide (dNTP) precursors (compared on page i.1: The Structure of Deoxyribonucleic acid). In addition, RNAs incorporate uracil (U) nucleotides into RNA strands instead of the thymine (T) nucleotides used in DNA. RNA polymerases differ from Dna polymerases in that they exercise non require primers. With the assistance of transcription initiation factors, RNA polymerase locates the transcription start site of a cistron and begins synthesis of a new RNA strand from scratch past joining the two ribonucleotides that are complementary to the first two bases of the template strand.
Overview of the Stages of Transcription
The basic steps of transcription are initiation, elongation, and termination. Here we tin identify several of the Dna sequences that characterize a gene. The promoter is the binding site for RNA polymerase. Information technology unremarkably lies v' to, or upstream of the transcription showtime site. Bounden of the RNA polymerase positions the enzyme to almost the transcription offset site, where it will showtime unwinding the double helix and begin synthesizing new RNA. The transcribed gray Dna region in each of the 3 panels are the transcription unit of the factor. Termination sites are typically 3' to, or downstream from the transcribed region of the gene. By convention, upstream refers to DNA 5' to a given reference point on the Dna (e.m., the transcription start-site of a cistron). Downstream then, refers to Deoxyribonucleic acid 3' to a given reference point on the DNA.
RNA polymerase
Building an RNA strand is very like to building a DNA strand. This is not surprising, knowing that DNA and RNA are very similar molecules. What enzyme carries out transcription? Transcription is catalyzed by the enzyme RNA Polymerase. "RNA polymerase" is a general term for an enzyme that makes RNA. At that place are many dissimilar RNA polymerases.

Like DNA polymerases, RNA polymerases synthesize new strands only in the v' to 3' direction, but because they are making RNA, they use ribonucleotides (i.e., RNA nucleotides) rather than deoxyribonucleotides. Ribonucleotides are joined in exactly the same way as deoxyribonucleotides, which is to say that the three'OH of the final nucleotide on the growing chain is joined to the v' phosphate on the incoming nucleotide.
One important deviation between DNA polymerases and RNA polymerases is that the latter do not require a primer to start making RNA. One time RNA polymerases are in the right place to start copying DNA, they merely begin making RNA by stringing together RNA nucleotides complementary to the Dna template.
This, of course, brings us to an obvious question- how exercise RNA polymerases "know" where to beginning copying on the Dna. Unlike the situation in replication, where every nucleotide of the parental Dna must somewhen be copied, transcription, as nosotros accept already noted, but copies selected genes into RNA at any given time.What indicates to an RNA polymerase where to start copying Dna to make a transcript? Signals in Dna indicate to RNA polymerase where information technology should start (and end) transcription. These signals are special sequences in Deoxyribonucleic acid that are recognized by the RNA polymerase or by proteins that help RNA polymerase determine where information technology should bind the DNA to offset transcription. A Dna sequence at which the RNA polymerase binds to kickoff transcription is called a promoter.
A promoter is by and large situated upstream of the gene that it controls. What this means is that on the DNA strand that the gene is on, the promoter sequence is "before" the gene. Call up that, by convention, DNA sequences are read from 5' to 3'. So the promoter lies 5' to the start point of transcription.
Likewise find that the promoter is said to "command" the gene information technology is associated with. This is considering expression of the gene is dependent on the binding of RNA polymerase to the promoter sequence to begin transcription. If the RNA polymerase and its helper proteins do not bind the promoter, the gene cannot be transcribed and it volition therefore, not exist expressed.

What is special near a promoter sequence? In an effort to answer this question, scientists looked at many genes and their surrounding sequences. Information technology makes sense that because the same RNA polymerase has to demark to many different promoters, the promoters should have some similarities in their sequences. Sure plenty, common sequence patterns were seen to exist present in many promoters. We will offset take a await at prokaryotic promoters. When prokaryotic genes were examined, the following features commonly emerged:
- A transcription starting time site (this the base in the Deoxyribonucleic acid across from which the first RNA nucleotide is paired).
- A -10 sequence: this is a half dozen bp region centered about x bp upstream of the start site. The consensus sequence at this position is TATAAT. In other words, if you count back from the transcription start site, which by convention, is called the +ane, the sequence found at -10 in the majority of promoters studied is TATAAT).
- A -35 sequence: this is a sequence at near 35 basepairs upstream from the first of transcription. The consensus sequence at this position is TTGACA.
What is the significance of these sequences? It turns out that the sequences at -x and -35 are recognized and bound by a subunit of prokaryotic RNA polymerase before transcription tin begin.
The RNA polymerase of Due east. coli, for instance, has a subunit called the sigma (σ) subunit (or sigma factor) in add-on to the cadre polymerase, which is the part of the enzyme that actually makes RNA. Together, the sigma subunit and core polymerase make upwardly what is termed the RNA polymerase holoenzyme. The sigma subunit of the polymerase can recognize and bind to the -10 and -35 sequences in the promoter, thus positioning the RNA polymerase at the correct place to initiate transcription. Once transcription begins, the core polymerase and the sigma subunit separate, with the core polymerase continuing RNA synthesis and the sigma subunit wandering off to escort some other core polymerase molecule to a promoter. The sigma subunit can exist idea of as a sort of usher that leads the polymerase to its "seat" on the promoter.
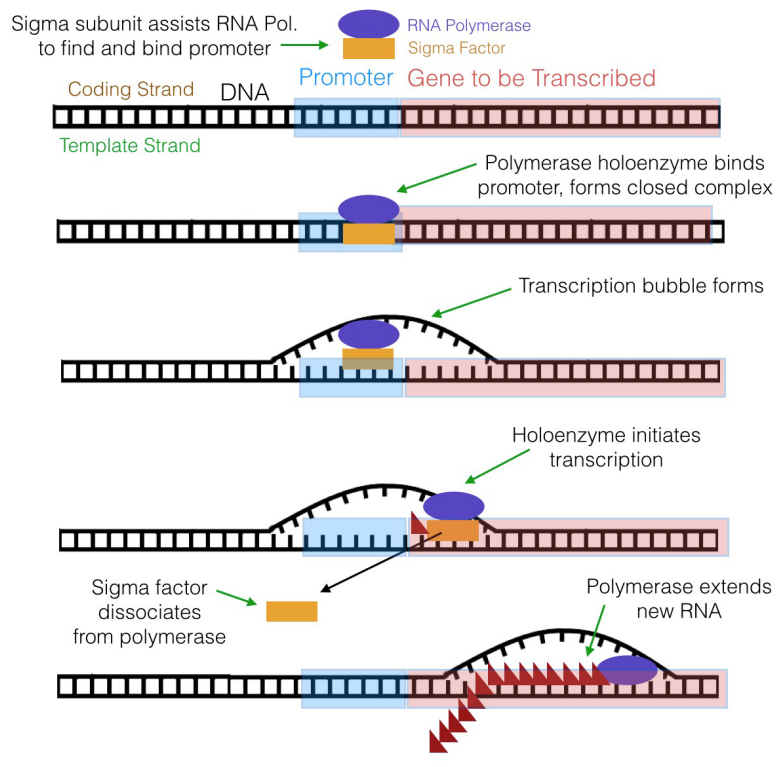
Equally already mentioned, an RNA concatenation, complementary to the Dna template, is built by the RNA polymerase by the joining of the v' phosphate of an incoming ribonucleotide to the 3'OH on the terminal nucleotide of the growing RNA strand. How does the polymerase know where to stop? A sequence of nucleotides chosen the terminator is the betoken to the RNA polymerase to stop transcription and dissociate from the template.
Although the process of RNA synthesis is the same in eukaryotes as in prokaryotes, there are some additional issues to keep in mind in eukaryotes. Ane is that in eukaryotes, the DNA template exists equally chromatin, where the DNA is tightly associated with histones and other proteins. The "packaging" of the Dna must therefore exist opened up to allow the RNA polymerase access to the template in the region to be transcribed.
A 2nd divergence is that eukaryotes have multiple RNA polymerases, not one every bit in bacterial cells. The different polymerases transcribe different genes. For example, RNA polymerase I transcribes the ribosomal RNA genes, while RNA polymerase III copies tRNA genes. The RNA polymerase we will focus on most is RNA polymerase 2, which transcribes protein-coding genes to brand mRNAs.
All 3 eukaryotic RNA polymerases demand additional proteins to help them get transcription started. In prokaryotes, RNA polymerase by itself tin can initiate transcription (remember that the sigma subunit is a subunit of the prokaryotic RNA polymerase). The boosted proteins needed by eukaryotic RNA polymerases are referred to as transcription factors.
Finally, in eukaryotic cells, transcription is separated in space and time from translation. Transcription happens in the nucleus, and the mRNAs produced are processed further before they are sent into the cytoplasm. Poly peptide synthesis (translation) happens in the cytoplasm. In prokaryotic cells, mRNAs tin can be translated as they are coming off the Deoxyribonucleic acid template, and because there is no nucleus, transcription and protein synthesis occur in a single cellular compartment.
Like genes in prokaryotes, eukaryotic genes also accept promoters. Eukaryotic promoters commonly have a TATA box, a sequence about 25 base pairs upstream of the start of transcription that is recognized and spring by proteins that help the RNA polymerase to position itself correctly to begin transcription. (Some eukaryotic promoters lack TATA boxes, and have, instead, other recognition sequences to assist the RNA polymerase discover the spot on the DNA where it spot on the Deoxyribonucleic acid where it binds and initiates transcription.)
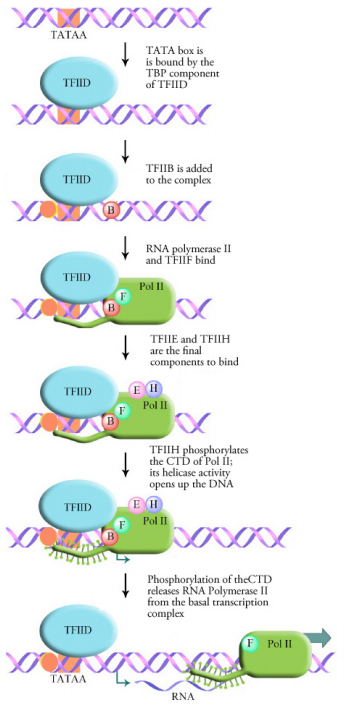
We noted before that eukaryotic RNA polymerases demand additional proteins to bind promoters and beginning transcription. What are these additional proteins that are needed to starting time transcription? General transcription factors are proteins that help eukaryotic RNA polymerases find transcription start sites and initiate RNA synthesis. We will focus on the transcription factors that assist RNA polymerase II. These transcription factors are named TFIIA, TFIIB and so on (TF= transcription factor, II=RNA polymerase 2, and the letters distinguish individual transcription factors).
Transcription in eukaryotes requires the general transcription factors and the RNA polymerase to course a circuitous at the TATA box called the basal transcription complex or transcription initiation complex. This is the minimum requirement for any gene to be transcribed. The starting time step in the formation of this complex is the binding of the TATA box by a transcription factor called the TATA Binding Poly peptide or TBP. Binding of the TBP causes the DNA to bend at this spot and take on a construction that is suitable for the binding of additional transcription factors and RNA polymerase. As shown in the figure at left, a number of different general transcription factors, together with RNA polymerase (Politico Two) form a complex at the TATA box.
The final footstep in the assembly of the basal transcription complex is the binding of a general transcription factor chosen TFIIH. TFIIH is a multifunctional protein that has helicase activeness (i.east., it is capable of opening up a DNA double helix) every bit well every bit kinase activeness. The kinase activity of TFIIH adds a phosphate onto the C-terminal domain (CTD) of the RNA polymerase. This phosphorylation appears to be the signal that releases the RNA polymerase from the basal transcription complex and allows it to move forward and begin transcription.
Either Dna strand can be a template
The promoter is the sequence of DNA that encodes the data almost where to begin transcription for each gene. Depending on the promoter, either strand of Dna can be used as the template strand.
Watch this video to meet how either strand of DNA can be used as a template for different genes on the same chromosome.
template vs. non-template strands summary
- The template strand is the one that RNA polymerase uses as the basis to build the RNA. This strand is also called the non-coding strand or the antisense strand.
- The non-template strand has the identical sequence of the RNA (except for the substituion of U for T). This strand is also called the coding strand or sense strand.
Major Types of Cellular RNA
Cells make several different kinds of RNA:
- mRNAs that code for proteins
- rRNAS that course part of ribosomes
- tRNAs that serve every bit adaptors betwixt mRNA and amino acids during translation
- MicroRNAs that regulate gene expression
- Other pocket-sized RNAs that take a variety of functions.
Contributors and Attributions
Source: https://bio.libretexts.org/Courses/University_of_Arkansas_Little_Rock/Genetics_BIOL3300_%28Fall_2021%29/Genetics_Textbook/02:_Central_Dogma/2.01:_Overview_of_Transcription
0 Response to "What Direction Does Rna Polymerase Read Dna"
Postar um comentário